7.8.15. Offline reweighting#
Introduction#
The majority of physics analyses at Belle II rely on simulation for setting up the selection cuts for the process of interest.
The selection on simulated data (MC) will often result in a different efficiency if the same selection is applied on the real experimental data,
hence data/MC corrections are necessary.
The data/MC tables can be applied online, i.e. during basf2 processing, or offline, on the resulting analysis ntuples.
One can find the latest approved correction tables for offline reweighting in the following Git repository.
These data/MC corrections often depend on particle properties, so they are typically provided in bins of kinematics, channel IDs, or other properties.
The add_weights_to_dataframe helper function can add the data/MC corrections to the ntuples according to the defined binning
and it can simplify the uncertainty propagation.
Warning
The add_weights_to_dataframe helper function in basf2 will soon be deprecated. A seperate sysvar python package will be recommended instead.
Charged Particle Identification corrections#
Typically, the Particle IDentification (PID) correction tables are provided in bins of momentum, polar angle and charge of a particle
and for a particular PID selection cut.
The offline PID tables are provided as files in CSV format. They can be easily loaded as a DataFrame in a Jupyter Notebook or in a python script
via the pandas.from_csv() function.
Each charged final state particle can have corresponding efficiency and fake rate correction tables.
For the offline reweighting one needs to specify the reconstructed and generated PDG codes for each table, e.g. the electron efficiency
table is (11, 11) and the pion to electron fake rate is (11, 211).
The user ntuple may contain variables for a mother particle and several daughter particles and
the daughter particles have their own unique prefix, so here we expect that all necessary variables for the reweighting have the same prefix.
The PID selection thresholds should be specified per particle to select correct entries from the PID correction tables.
The following code snippet provides an example of reweighting particles with prefixes lepminus_ and lepplus_ in the user_df DataFrame:
from sysvar import add_weights_to_dataframe
tables = {(11, 11): efftable_e,
(11, 211): faketable_pi_e,
(11, 321): faketable_K_e}
thresholds = {11: ('pidChargedBDTScore_e', 0.9)}
add_weights_to_dataframe('lepminus_',
user_df,
systematic='custom_PID',
custom_tables=tables,
custom_thresholds=thresholds)
add_weights_to_dataframe('lepplus_',
user_df,
systematic='custom_PID',
custom_tables=tables,
custom_thresholds=thresholds)
Warning
Please make sure that the correction tables and the ntuple are compatible before application of the weights!
In the example above, it is expected that pidChargedBDTScore_e > 0.9 cut has been already applied on user_df.
By default, the add_weights_to_dataframe function calls will add lepminus_Weight and lepplus_Weight
columns to the user_df and they will add additional columns of lepminus_Weight_0 to lepminus_Weight_99 and
lepplus_Weight_0 to lepplus_Weight_99, which are the variations of the nominal weights according to the total PID uncertainty.
The systematic uncertainties can be correlated by providing the same seed value to the particles
by providing the same sys_seed argument value to the add_weights_to_dataframe method calls.
One can switch off the weight variations by providing the generate_variations=False flag to the add_weights_to_dataframe function,
and in this case only data_MC_ratio and the corresponding statistical and systematic uncertainty columns will be added to the ntuples
with corresponding particle prefix.
The coverage plots can be shown by providing the show_plots=True argument.
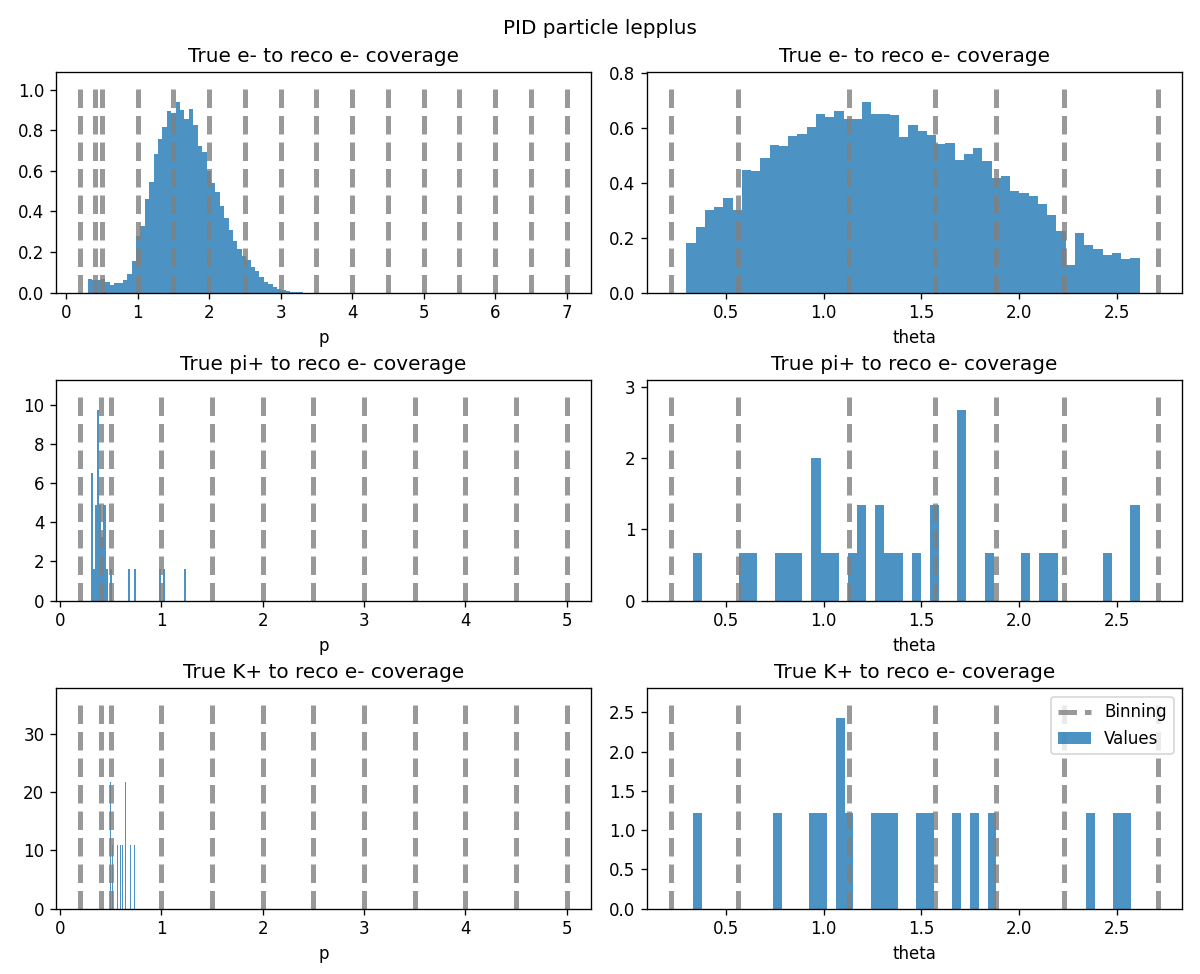
This plot shows the distributions of momentum and theta of the lepplus particle split by mcPDG values,
and the binning of the corresponding correction tables.
The entries that are not covered by the correction tables will assigned a NaN value, which is replaced by default with 1.0 and
the replaced value can be regulated via fillna argument.
Full Event Interpretation corrections#
Similarly to PID weights, the FEI tables are provided as files in CSV format. The corrections are binned in FEI decay mode ID and they are provided for several thresholds of FEI probability. Because of the high cross-feed rates between the FEI modes, a covariance matrix must be provided.
from sysvar import add_weights_to_dataframe
add_weights_to_dataframe('Btag',
user_df,
systematic='custom_FEI',
custom_tables=table,
custom_thresholds=0.01,
cov_matrix=cov,
variable_aliases={'dec_mode': 'Btag_mode'},
show_plots=True)
In the snippet above, the reweighting is performed for Btag particle in user_df DataFrame.
The FEI correction table is loaded as table DataFrame via the pandas.read_csv() function and the covariance matrix cov
is loaded as a separate numpy.ndarray via the numpy.load() (.npy files) or the numpy.loadtxt() (.tsv files) method.
The add_weights_to_dataframe function expects that the FEI decay mode ID is provided
in the {prefix}_dec_mode column of the user_df, but if this is not the case, one can provide a variable alias
to point the object to the existing column.
Modules and functions#
This section provides the documentation for the classes that are used for offline reweighting.
- sysvar.add_weights_to_dataframe(prefix: str, df: DataFrame, systematic: str, custom_tables: dict = None, custom_thresholds: dict = None, **kw_args) DataFrame[source]#
Helper method that adds weights to a dataframe.
- Parameters:
prefix (str) – Prefix for the new columns.
df (pandas.DataFrame) – Dataframe containing the analysis ntuple.
systematic (str) – Type of the systematic corrections, options: “custom_PID” and “custom_FEI”.
MC_production (str) – Name of the MC production.
custom_tables (dict) – Dictionary containing the custom efficiency weights.
custom_thresholds (dict) – Dictionary containing the custom thresholds for the custom efficiency weights.
n_variations (int) – Number of variations to generate.
generate_variations (bool) – When true generate weight variations.
weight_name (str) – Name of the weight column.
show_plots (bool) – When true show the coverage plots.
variable_aliases (dict) – Dictionary containing variable aliases.
cov_matrix (numpy.ndarray) – Covariance matrix for the custom efficiency weights.
fillna (int) – Value to fill NaN values with.
sys_seed (int) – Seed for the systematic variations.
syscorr (bool) – When true assume systematics are 100% correlated defaults to true.
**kw_args – Additional arguments for the Reweighter class.